
The vaginal microbiota, high-risk human papillomavirus infection, and cervical cytology: results from a population-based study
Introduction
Annually, approximately 530,000 women globally develop invasive cervical cancer (1-4). Persistent infection with high-risk human papillomavirus (HR HPV) is strongly associated with the development of abnormal cervical cytology and progression to cervical cancer over many years (5). Specifically, HR HPV types 16, 18, and 45 account for the majority of cases, with HPV-16 alone accounting 50–55% of cervical cancer cases (6). Factors associated with viral persistence and pathways to progression to cervical cancer are not well understood. Natural history observational studies have shown that 10% of HPV infections remain persistent, and nearly half of the persistent infections progress to cervical intraepithelial neoplasia (CIN) grade 3; with only 20% of CIN 3 progressing to cervical cancer in five years (7). The reasons for progression and persistence are not known.
Epidemiologic studies have demonstrated associations between bacterial vaginosis (BV), as diagnosed by Nugent’s criteria (i.e., scoring based on light-microscopic examination of vaginal discharge), and persistence of HPV infection and disease severity (8-10). Emerging evidence from next generation sequencing studies suggest that specific community types of vaginal microbiota may be temporally associated with HPV persistence and is one factor that could provide insight into cervical cancer progression—either as a risk marker or risk mediator (11,12).
Culture-independent methods such as 16S rRNA gene-encoding amplicon sequencing studies have confirmed prior work demonstrating that vaginal microbiota are often dominated by a single species of Lactobacillus. The hypothesized function of the vaginal microbiota is to provide colonization resistance to pathogens by lowering the pH of the vagina via lactic acid production (13-16), or by directly interacting with the host immune system (17). Vaginal microbial community state types have been described based on the absence or presence of the predominant Lactobacillus type, and absence of Lactobacillus spp. has been associated with adverse health outcomes (16). Diverse, poly-microbial vaginal communities marked by Lactobacillus depletion and abundant anaerobes have been associated with BV, adverse reproductive outcomes such as sexually transmitted infections and HIV acquisition, as well as preterm birth (18). It is also possible that vaginal microbiota may biologically mediate the associations between persistent HPV infections and risk factors for cervical cancer.
Our primary objective was to characterize the vaginal microbiota using next generation sequencing from a stratified random sample of women from a population-based study in Appalachia—a region that has the highest annual rate of cervical cancer mortality in the United States (19), and compare compositional differences among women with abnormal cervical cytology regardless of any HPV infection (CC), women with HR HPV+ infection only without cytologic abnormality (HPV+), and women without cytologic abnormalities (negative for intraepithelial lesion or malignancy) or HPV infection (NILM/HPV−), as well as demographic, behavioral, and clinical risk factors.
Methods
We analyzed residual liquid cytology samples from women in the Community Access, Resources and Education (CARE): Project 3 study across 16 clinics in southeast Ohio and West Virginia. Details of the main study have been published elsewhere (20). Eligible women were ≥18 years, resided in Ohio Appalachian county, were not pregnant, seen in a participating clinic, and had no history of hysterectomy or invasive cervical cancer. Data on antibiotic use were not available from the Community Access, Resources and Education: Project 3 study. From the 308 women who had complete biological and clinical data in the CARE 3 cohort, we randomly chose women within three strata: (I) 109 women with abnormal cervical cytology [i.e., the majority were atypical squamous cells of undetermined significance (n=55 ASC) and low-grade squamous intraepithelial lesions (n=45 LGSIL) while n=6 were high-grade squamous intraepithelial lesions and n=3 were atypical glandular cells]; (II) 110 HR HPV+ women; and (III) 89 NILM/HPV− women. Among the women with abnormal cervical cytology (n=109), 80 were HPV+, the majority of which were positive for a high-risk type of HPV (n=61). Selection was based on stratification by outcome then random selection within each stratum. This study was deemed not regulated by the University of Michigan Institutional Review Board since it included de-identified previously collected data (Table 1).
Table 1
| Characteristics | Entire population (N=308) | Abnormal cervical cytology (N=109), (35.4%) | HR HPV+ (N=110), (35.7%) | NILM/HPV− (N=89), (28.9%) | P# |
|---|---|---|---|---|---|
| Age, median [IQR] | 26 [21–39] | 24 [21–34] | 24 [21–34] | 35 [25–48] | <0.001* |
| Race/ethnicity, N (%) | |||||
| White | 291 (94.5) | 105 (96.3) | 103 (93.6) | 83 (93.3) | 0.22 |
| Black or African American | 6 (1.9) | 0 (0) | 4 (3.6) | 2 (2.2) | |
| American Indian or Alaskan Native | 1 (0.3) | 1 (0.9) | 0 (0) | 0 (0) | |
| Asian | 5 (1.6) | 3 (2.8) | 1 (0.9) | 1 (1.1) | |
| Hispanic | 1 (0.3) | 1 (0.9) | 0 (0) | 0 (0) | 0.64 |
| Completed high school, N (%) | 271 (88.0) | 97 (89.0) | 96 (87.3) | 78 (87.6) | 0.95 |
| Smoking, N (%) | |||||
| Smoked ≥100 lifetime cigarettes | 188 (61.0) | 73 (67.0) | 71 (64.5) | 44 (49.4) | 0.06 |
| Current smoker | 143 (46.4) | 59 (54.1) | 57 (51.8) | 27 (30.3) | 0.01* |
| Alcohol consumption, N (%) | |||||
| Light/moderate | 108 (35.1) | 37 (33.9) | 38 (34.5) | 33 (37.1) | 0.08 |
| Heavy | 92 (29.9) | 41 (37.6) | 34 (30.9) | 17 (19.1) | |
| Married, N (%) | 86 (27.9) | 24 (22.0) | 23 (20.9) | 39 (43.8) | <0.001* |
| 2+ male partners in last year, N (%) | 92 (29.9) | 45 (41.3) | 37 (33.6) | 10 (11.2) | <0.001* |
| Contraception, N (%) | |||||
| History of hormonal method or copper IUD† | 275 (89.3) | 102 (93.6) | 101 (91.8) | 72 (80.9) | 0.04* |
| History of non-hormonal method‡ | 122 (39.6) | 45 (41.3) | 39 (35.5) | 38 (42.7) | 0.42 |
| Occasional current condom use | 64 (20.8) | 28 (25.7) | 25 (22.7) | 11 (12.4) | 0.02* |
| Regular current condom use | 52 (16.9) | 17 (15.6) | 23 (20.9) | 12 (13.5) | |
| Menopause, N (%) | 33 (10.7) | 8 (7.3) | 8 (7.3) | 17 (19.1) | 0.01* |
| HPV-related history, N (%) | |||||
| History of abnormal pap | 146 (47.4) | 64 (58.7) | 52 (47.3) | 30 (33.7) | 0.001* |
| History of warts, condyloma, or HPV | 41 (13.3) | 19 (17.4) | 17 (15.5) | 5 (5.6) | 0.05 |
| Vaginal microbiota, N (%) | |||||
| CST 1—L. crispatus dominated | 56 (18.2) | 19 (17.4) | 21 (19.1) | 16 (18.0) | 0.04* |
| CST 2—L. iners dominated | 116 (37.7) | 35 (32.1) | 36 (32.7) | 45 (50.6) | |
| CST 3—Diverse | 136 (44.2) | 55 (50.5) | 53 (48.2) | 28 (31.5) | |
| L. gasseri present | 205 (66.6) | 74 (67.9) | 62 (56.4) | 69 (77.5) | 0.01* |
#, Kruskal-Wallis nonparametric ANOVAs were used to compare continuous variables between women with abnormal cervical cytology, women with HR-HPV+, and women with NILM/HPV−. Fisher’s exact tests were used to compare race and Hispanic between women with abnormal cervical cytology, women with HR-HPV, and women with NILM/HPV−. Chi squared tests were used to compare all other categorical variables between women with abnormal cervical cytology, women with HR-HPV+, and women with NILM/HPV− screens. †, Methods included contraceptive pill, patch, injections, vaginal ring, and hormonal IUD. ‡, Methods included contraceptive sponge, diaphragm, spermicide, withdrawal, and natural family planning. *, indicates significance at P<0.05 level. N, number; HR HPV+, high risk human papillomavirus; IQR, interquartile range; IUD, intrauterine device; pap, pap smear; HPV, human papillomavirus; CST community state type.
Vaginal microbiota was characterized using 16S rRNA gene amplicon sequencing of the V4 region on the Illumina MiSeq platform. Nucleic acids were isolated using the MagAttract PowerMicrobiome DNA/RNA kit (Qiagen). Amplification of the V4 region of the 16S rRNA gene was done by standard PCR on 3 or 7 µL of DNA as described previously (21) or, if needed, 3 µL of DNA was amplified by touchdown PCR [1×(2 min at 95 °C), 20×(20 s at 95 °C, 15 s at annealing temperature, starts at 60 °C, decreases 0.3 °C/cycle), 5 min at 72 °C, 20×(20 s at 95 °C, 15 s at 55 °C, 5 min at 72 °C), 1×(10 min at 72 °C)]. All data from samples with <1,000 sequences were discarded, and the UCHIME algorithm was used to detect chimeric sequences (22). Quality scores were generated following procedures in the MiSeq SOP (23). Specifically, after alignment of the paired end read length of 250 base pairs and identification of positions where the two reads disagree, for any sequence that has a base and the other a gap, the quality score of the base must be over 25 to be considered real. If both sequences have a base at that position, then one of the bases must have a quality score of ≥6 points more than the other. If it is less than 6 points better, then the consensus base is set to an N. Sequence files were deposited in the NCBI Sequence Read Archive (PRJNA622998).
Sequences were processed and analyzed using mothur (v.1.39.5) and MiSeq SOP (23,24). After alignment to Silva {release 12 [8]} (25,26), sequences were assigned to operational taxonomic units (OTUs) based on 97% sequence similarity using the OptiClust method (27), and taxonomically classified using a modified version of the Ribosomal Database Project (RDP) (version 16) (28) according to previously published methods (23). After alignment and classification, we used the Remove.lineage tool to remove non-bacterial sequences.
We calculated Shannon and Inverse Simpson metrics to examine sample richness and alpha diversity and plotted them by outcome. θYC distances between samples were calculated, a measure of compositional difference between communities, based on the relative abundances OTUs that are shared between two communities, and those that are unique to either community (29). Community state types (CSTs) were identified using partitioning around the medoid clustering based on θYC distances. Linear discriminant analysis effect size (LEfSe) (30), which uses statistical and biological significance to determine features that are differential between groups, and rank those features according to effect size, was used to identify OTUs that were significantly differentially abundant across the three outcome groups. PCoA plots of thetayc distances and AMOVA were used to plot and test for differences between abnormal cytology v HR HPV+, abnormal cytology v NILM/HPV−, and HR HPV+ v NILM/HPV−, respectively.
Kruskal-Wallis, Chi squared tests, and Fisher’s exact tests where appropriate, were used to test for associations between subject characteristics and strata. Multinomial logistic regression models were used to test for associations between CC and HR HPV+ status, respectively, compared to controls and vaginal microbiota exposures, which included both vaginal CSTs as well as the presence of L. gasseri. Potential confounders were selected based on a priori knowledge and bivariate associations between subject characteristics with health status and vaginal microbiota community types, respectively.
Results
The sample of 308 women was comprised of 94.5% non-Hispanic White women from Appalachia, with a mean age of 26 (IQR =21–39) years. More than 88% had at least a high school education while 29% were married. Overall, 46% of the sample currently smoked, and 61% reported a history of smoking at least 100 cigarettes. Nearly a third reported heavy (35.1%), or moderate (29.9%), alcohol consumption (Table 1).
Women who were HR HPV+ (n=110) without cytological abnormality were younger on average than NILM/HPV− women [mean =24 (IQR =21–34) years] compared to 35 years (IQR =25–48 years, P<0.001). Current smoking, single marital status, and having at least 2 partners in the last year were more common among women with HR HPV+ women compared to NILM/HPV− women (Table 1). Women with abnormal cervical cytology (n=128) also were younger on average [mean =25 years (IQR =20–36) years] than NILM/HPV− women (Table 1). Current smoking, single marital status, and having at least 2 partners in the last year were more common among women with abnormal cervical cytology compared to NILM/HPV− women (Table 1). There were also differences in contraception methods across the three strata; a history of using either hormonal contraception or copper IUDs and only occasional use of condoms was more common among women with abnormal cervical cytology and HR HPV+ compared to NILM/HPV− women (Table 1).
Samples from 308 women had at least 1,000 sequences after processing and were included in the analysis. There was an average of 27,418 (±12,921 SD) sequences per sample (Table S1). We observed three vaginal microbiota CSTs in this sample of women from Appalachia. Mean relative abundances of major taxa by vaginal microbiota CST are shown in Figure 1. The most common CST 3 was a diverse, poly-microbial state characterized by higher Gardnerella vaginalis relative abundance prevalent in 44% of women, followed by CST 2 which was L. iners dominant prevalent in 37.7% while CST 1, L. crispatus dominant, was present in only 18.2% of women.
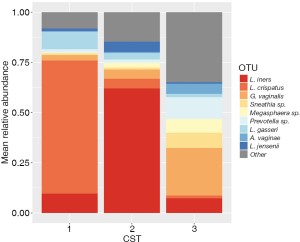
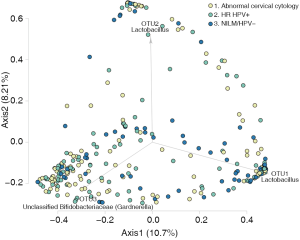
Women with abnormal cervical cytology or HR HPV+ women were more likely to have CST 3 compared to NILM/HPV− women whose communities were more likely to be Lactobacillus spp. dominant (P=0.04). Women across all outcome groups were similarly likely to have L. crispatus dominated communities (CST 1). NILM/HPV- women had a higher prevalence of L. iners dominated communities (CST 2) than women with abnormal cervical cytology and HR HPV+ respectively (P=0.04, Table 1). Major taxa relative abundances by health status are shown in the heat map in (Figure 2). There were not striking differences in richness or alpha diversity across the three outcome groups (Figures 3 and 4). The relative abundance of L. gasseri was significantly greater among NILM/HPV− women than among women with abnormal cervical cytology or HR HPV+ by LEfSe (LDA =4.17; P=0.0009) (Figure S1). Differences in beta diversity were detected using AMOVA on thetayc distances overall across the three outcomes (P=0.04) as well as between abnormal cytology and NILM/HPV (P=0.02) and between HR HPV+ and NILM/HPV− (P=0.017) (Figure 5).
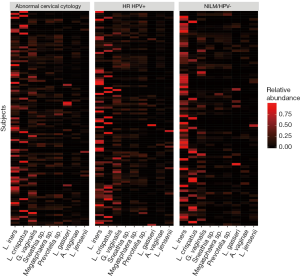
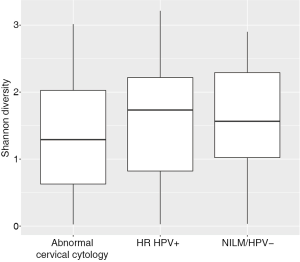
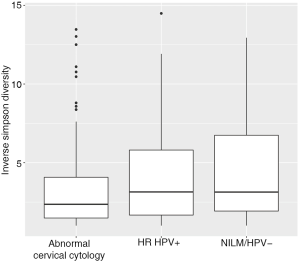
Based on a priori evidence of potential confounding, we included age, race, and current smoking in adjusted models along with ≥2 male partners in the last year which was associated with both CST (not shown) and outcome (Table 1). In these adjusted multinomial logistic regression models, the associations between abnormal cervical cytology status and CST 3 and between HR HPV+ status and CST 3 we had observed using chi-square tests (Table 1) were no longer significant [abnormal cervical cytology (OR =1.63; 95% CI: 0.66–4.03) and HR HPV+ (OR =1.53; 95% CI: 0.62–3.76) respectively) (Table 2). L. iners dominated communities (CST 2) were also not significantly associated with abnormal cervical cytology or HR HPV+ [abnormal cervical cytology (adjusted OR =0.67; 95% CI: 0.28–1.59) and HR HPV+ (adjusted OR =0.67; 95% CI: 0.29–1.57) respectively (Table 2)].
Table 2
| Variable | Crude (N=308) | Adjusted (N=284) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abnormal cervical cytology† | HR HPV+† | Abnormal cervical cytology† | HR HPV+† | ||||||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||
| Age | – | – | – | – | 0.96* | 0.94–0.99* | 0.96* | 0.93–0.98* | |||
| Community state type | |||||||||||
| CST 1, L. crispatus dominated | Referent | Referent | |||||||||
| CST 2, L. iners dominated | 0.65 | 0.29–1.46 | 0.61 | 0.28–1.34 | 0.67 | 0.28–1.59 | 0.67 | 0.29–1.57 | |||
| CST 3, diverse | 1.65 | 0.74–3.70 | 1.44 | 0.65–3.20 | 1.63 | 0.66–4.03 | 1.53 | 0.62–3.76 | |||
| Race | |||||||||||
| African American/American Indian/Alaskan Native/Asian | Referent | Referent | |||||||||
| White race | – | – | – | – | 1.08 | 0.15–7.53 | 0.83 | 0.13–5.26 | |||
| Tobacco status | |||||||||||
| Non smoker | Referent | Referent | |||||||||
| Current smoker | – | – | – | – | 1.75 | 0.89–3.46 | 1.69 | 0.69–4.16 | |||
| Sexual partners | |||||||||||
| 1 or fewer male partners in last year | Referent | Referent | |||||||||
| 2+ male partners in last year | – | – | – | – | 3.17* | 1.30–7.74* | 2.34 | 0.84–6.52 | |||
†, reference group was NILM/HPV− women. *, indicates significance at P<0.05. N, number, abnormal cervical cytology; HR HPV, high risk human papillomavirus; OR, odds ratio; CI, confidence interval; CST, community state type.
When we modeled presence of L. gasseri relative abundance in relation to the outcomes with crude and adjusted multinomial logistic regression models, L. gasseri was significantly inversely associated with HR HPV+ status (OR =0.37; 95% CI: 0.20–0.70) but did not reach significance for abnormal cervical cytology compared to NILM/HPV− (OR =0.61; 95% CI: 0.32–1.16) (Table 3). After adjustment for age, white race, current smoking, 2+ male partners in the last year, and current condom use, age was significantly inversely associated with both HR HPV+ status and abnormal cervical cytology, current smoking was significantly positively associated with HR HPV+ status, ≥2 male partners in the last year was significantly positively associated with both HPV and abnormal cervical cytology, and current condom use was significantly inversely associated with HPV (Table 3). Presence of L. gasseri was barely no longer significantly associated with either HPV (adjusted OR =0.50; 95% CI: 0.25–1.02) or cervical cytology compared to NILM/HPV− (adjusted OR =0.88; 95% CI: 0.42–1.83).
Table 3
| Variable | Univariate (N=308) | Multivariate (N=257) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abnormal cervical cytology† | HR HPV+† | Abnormal cervical cytology† | HR HPV+† | ||||||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||
| L. gasseri present ‡ | 0.61 | 0.32–1.16 | 0.37* | 0.20–0.70* | 0.88 | 0.42–1.83 | 0.50 | 0.25–1.02 | |||
| Age | – | – | – | – | 0.97* | 0.94–1.00* | 0.97* | 0.94–1.00* | |||
| White race§ | – | – | – | – | 0.70 | 0.07–7.42 | 0.63 | 0.06–6.24 | |||
| Current smoker¶ | – | – | – | – | 1.96 | 1.00–3.87 | 2.03* | 1.03–4.01* | |||
| 2+ male partners in last year†† | – | – | – | – | 3.77* | 1.56–9.09* | 2.46* | 1.01–6.02* | |||
| Current condom use (ordinal)# | – | – | – | – | 0.88 | 0.62–1.24 | 0.69* | 0.49–0.96* | |||
†, reference was NILM/HPV− women. ‡, reference was absence of L. gasseri. §, reference group was black or African American, American Indian or Alaskan Native, and Asian subjects. ¶, reference group was current non-smokers. ††, reference group was subjects with one or fewer male partners in the last year. #, levels from low to high were: no current partner (reference level), regular current condom use, occasional current condom use, no current condom use. *, indicates significance at P<0.05. N, number; HR HPV, high risk human papillomavirus; OR, odds ratio; CI, confidence interval; pap, pap smear.
Discussion
In this cohort of women from Appalachia, the vaginal microbiota of women with abnormal cervical cytology as well as women with HR HPV+ alone compared to women with neither condition was characterized by a diverse community type with high relative abundance of G. vaginalis and reduced relative abundance of Lactobacillus. However, after adjustment, these differences were attenuated. Several factors may explain our observed results. First, the most common vaginal community type in our sample overall was a diverse community type with diminished Lactobacillus. While clustering methods differ, Lactobacillus-depleted community types have been reported among 10–42% of women in other studies (31), depending on the population sampled while only a minority of women had a L. crispatus-dominant community in our study. It is thus possible that Lactobacillus depleted communities are prevalent among women of Appalachia as compositional differences have been observed across other cohorts. For example, higher relative proportion of L. iners-dominance among Hispanic women compared to White cohorts have been demonstrated in other studies (16,32). It is unclear whether differences in vaginal community composition are due to increased prevalence of asymptomatic BV in these populations (33), increased environmental and behavioral risk factors, or underlying genetic risk.
Our results are fairly consistent with other recent studies that have characterized vaginal microbiota signals associated with HPV. Overall, both microscopy and culture-independent studies support a consistent, moderate association between non-Lactobacillus-dominated microbiota and HPV (34,35), and women with depleted Lactobacillus communities have been shown to have the slowest rates of HPV remission compared to women with L. crispatus-dominant CSTs (34). Across diverse cohorts, studies on Black South African (36), Chinese (37), and Korean (38,39) women have demonstrated similar findings associating HPV infection with a lower proportion of Lactobacillus spp. compared to HPV− women regardless of the proportion of Lactobacillus spp. dominance in the population. There appears to be some evidence that vaginal microbiota may be related to viral persistence (40), although few studies to date have been longitudinal. Women who are HPV+ also appear to differ from HPV− women in terms of several key vaginal metabolites, including amines, glutathione, and lipids (41). However, the majority of studies to date have not adequately adjusted for potential confounders, which may explain the attenuation of some of the effects we observed after adjustment for such factors.
Our results are also consistent with other recent studies that have used culture-independent methods to characterize vaginal microbiota signals associated with the development and severity of cervical cancer (11,42). Mitra et al. found increased vaginal microbiota diversity was associated with CIN disease severity among a sample of 169 women with pre-invasive and invasive disease and HPV− controls, as well as enrichment of key taxa differentially abundant among women with more advanced disease (43). Although their results suggested vaginal microbiota could potentially be used as a microbiological marker of clinically significant disease, there was no adjustment for potential confounders, which was a major limitation of their study. Recently, cancer biomarker signatures sampled cross-sectionally from the cervicovaginal microenvironment revealed patient metabolic profiles were driven by genital inflammation (44), HPV infection, and Lactobacillus spp. (45).
It is also possible that in some cases the vaginal microbiota serves as a biological mediator between clinical and demographic factors and cervical disease, but longitudinal studies will be needed to further explore. Vaginal microbiota may confer a range of health effects dependent on bacterial community composition that are protective of cervical disease. For example, L. crispatus-dominant communities are known to produce both isomers of lactic acid that render the environment inhospitable to pathogens (13-15), although interactions among host cervical cells, the microbiota, and metabolites are not yet well understood. Community shifts, which are common in non-L. crispatus-dominant communities, may lead to pro-inflammatory effects leading to tissue damage, genomic instability, and viral integration to ultimately promote development of cervical cancer. Independent of pH and lactate, in vitro studies indicate that Lactobacillus spp. exert cytotoxic effects on cervical tumor cells but not on normal cervical cells (46). However, it is equally possible that vaginal microbiota, in concert with HPV, can epigenetically impact the cervical cells, as well as the possibility that infection with HPV leads to perturbations in the vaginal microbiota. Questions that remain include whether a threshold effect exists for the both common taxa such as Lactobacillus, as well as that of more rare taxa with pathogenic potential.
Our study broadens the scope of the current literature by focusing on women from Appalachia, which has the highest annual rate of cervical cancer mortality in the United States (19). Our study has several strengths that add to this body of evidence including a large sample size from a population-based cohort, use of molecular methods of characterizing vaginal microbiota, adjustment of confounders, and the inclusion of a unique, high-risk sample. Our results extend the current understanding of altered vaginal microbiota among high-risk women by identifying community types and specific taxa with cervical disease.
The primary limitation of our study includes its cross-sectional design. Data on CIN staging and HPV persistence were thus not available. Given there is some heterogeneity with our study strata, we are likely underestimating the association between aberrant vaginal microbiota and cervical disease. Microbiota characterization using 16S gene amplicon sequencing may be biased against rare vaginal taxa or sequence variants with high pathogenic potential that may require deeper sequencing for better resolution or may be impacted by more limited vaginal data curation of reference databases. While our data cannot establish an etiologic role of the vaginal microbiota in CIN and HR HPV, it suggests that vaginal communities that are Lactobacillus-depleted may help to identify women at higher risk of HPV acquisition and persistence leading to abnormal cervical cytology—i.e., vaginal microbiota signatures may be risk markers even if they are not causally related to CIN. Further research is needed to study these potential mechanisms driving the observed associations between high-risk vaginal microbiota and CIN and HR HPV, especially in longitudinal studies of high-risk populations.
Our results suggest that compared women without cytologic abnormalities, the vaginal microbiota of women with abnormal cervical cytology or who were high-risk HPV+ were characterized by a diverse community with increased relative abundance of G. vaginalis and reduced relative abundance of L. gasseri. However, these differences were attenuated after adjustment and that other factors may be driving these associations. Given the invasiveness of current screening methods, further study of diverse cohorts is warranted given self-collected swabs and molecular based methods of identification may offer more attractive screening options for patients.
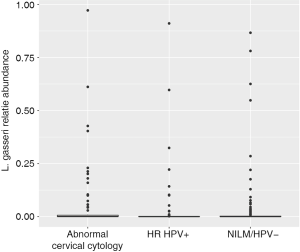
Table S1
| HPV status | Abnormal cervical cytology | Number of samples | Bacterial reads | Number of OTUs |
|---|---|---|---|---|
| High-risk | Yes | 61 | 1,656,906 | 2,855 |
| No | 110 | 3,017,918 | 4,931 | |
| Low-risk | Yes | 46 | 1,257,391 | 2,124 |
| No | 89 | 2,548,744 | 4,103 | |
| None | Yes | 2 | 54,396 | 92 |
| No | 0 |
OTU, operational taxonomic unit.
Acknowledgments
We would like to thank the University of Michigan Microbial Systems Molecular Biology Laboratory for DNA isolation and sequencing, as well as the CARE 3 study, and The Ohio State University for providing the samples and data.
Funding: This work was supported by grants from the National Cancer Institute (P50 CA105632, P30 CA016058, P30 CA046592) and the National Center for Advancing Translational Science (UL1TR001070).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Filipa Godoy-Vitorino) for the series “The Role of Microbiomes in the Development of HPV-cervical Cancer” published in Gynecology and Pelvic Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gpm.amegroups.org/article/view/10.21037/gpm-20-10/coif). The series “The Role of Microbiomes in the Development of HPV-cervical Cancer” was commissioned by the editorial office without any funding or sponsorship. MTR reports grants from National Cancer Institute, grants from National Center for Advancing Translational Science, during the conduct of the study. VBY reports other from Bio-K+ International, other from Vedanta Biosciences, Inc., other from Pantheryx, Inc., outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that the questions related to the accuracy and integrity of any part of the work are thoroughly investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was deemed not regulated by the University of Michigan Institutional Review Board since it included de-identified previously collected data (HUM00130947). Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- International Agency for Research on Cancer - World Health Organization, editor. Cervical Cancer. Estimated Incidence, Mortality and Prevalence Worldwide in 2012. GLOBOCAN; 2012; Lyon, France.
- Peto J, Gilham C, Fletcher O, et al. The cervical cancer epidemic that screening has prevented in the UK. Lancet 2004;364:249-56. [Crossref] [PubMed]
- U.S. Cancer Statistics Working Group. U.S. Cancer Statistics Data Visualizations Tool, based on November 2018 submission data (1999-2016). Centers for Disease Control and Prevention and National Cancer Institute, U.S. Department of Health and Human Services. 2019. Available online: www.cdc.gov/cancer/dataviz
- Harper DM, DeMars LR. HPV vaccines - A review of the first decade. Gynecol Oncol 2017;146:196-204. [Crossref] [PubMed]
- Bosch FX, Burchell AN, Schiffman M, et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine 2008;26:K1-16. [Crossref] [PubMed]
- Schiffman M, Rodriguez AC. Heterogeneity in CIN3 diagnosis. Lancet Oncol 2008;9:404-6. [Crossref] [PubMed]
- Gillet E, Meys JF, Verstraelen H, et al. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta-analysis. BMC Infect Dis 2011;11:10. [Crossref] [PubMed]
- Guo YL, You K, Qiao J, et al. Bacterial vaginosis is conducive to the persistence of HPV infection. Int J STD AIDS 2012;23:581-4. [Crossref] [PubMed]
- King CC, Jamieson DJ, Wiener J, et al. Bacterial vaginosis and the natural history of human papillomavirus. Infect Dis Obstet Gynecol 2011;2011:319460. [Crossref] [PubMed]
- Brusselaers N, Shrestha S, van de Wijgert J, et al. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: systematic review and meta-analysis. Am J Obstet Gynecol 2019;221:9-18.e8. [Crossref] [PubMed]
- van de Wijgert JHHM, Borgdorff H, Verhelst R, et al. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One 2014;9:e105998. [Crossref] [PubMed]
- Borges S, Silva J, Teixeira P. The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet 2014;289:479-89. [Crossref] [PubMed]
- Boskey ER, Cone RA, Whaley KJ, et al. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum Reprod 2001;16:1809-13. [Crossref] [PubMed]
- Hickey RJ, Zhou X, Pierson JD, et al. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res 2012;160:267-82. [Crossref] [PubMed]
- Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108:4680-7. [Crossref] [PubMed]
- Rose WA 2nd, McGowin CL, Spagnuolo RA, et al. Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PLoS One 2012;7:e32728. [Crossref] [PubMed]
- Leitich H, Bodner-Adler B, Brunbauer M, et al. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol 2003;189:139-47. [Crossref] [PubMed]
- Horner MJ, Altekruse SF, Zou Z, et al. U.S. geographic distribution of prevaccine era cervical cancer screening, incidence, stage, and mortality. Cancer Epidemiol Biomarkers Prev 2011;20:591-9. [Crossref] [PubMed]
- Reiter PL, Katz ML, Ruffin MT, et al. HPV prevalence among women from Appalachia: results from the CARE project. PLoS One 2013;8:e74276. [Crossref] [PubMed]
- Seekatz AM, Theriot CM, Molloy CT, et al. Fecal Microbiota Transplantation Eliminates Clostridium difficile in a Murine Model of Relapsing Disease. Infect Immun 2015;83:3838-46. [Crossref] [PubMed]
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. [Crossref] [PubMed]
- Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79:5112-20. [Crossref] [PubMed]
- Mothur. MiSeq SOP. 2019. Available online: https://www.mothur.org/wiki/MiSeq_SOP. Accessed January 31 2020.
- Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007;35:7188-96. [Crossref] [PubMed]
- Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 2009;4:e8230. [Crossref] [PubMed]
- Westcott SL, Schloss PD. OptiClust, an Improved Method for Assigning Amplicon-Based Sequence Data to Operational Taxonomic Units. mSphere 2017;2:e00073-17. [Crossref] [PubMed]
- Cole JR, Wang Q, Fish JA, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42:D633-42. [Crossref] [PubMed]
- Yue JC, Clayton MK. A Similarity Measure Based on Species Proportions. Comm Stat Theory Methods 2005;34:2123-31. [Crossref]
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [Crossref] [PubMed]
- Ravel J, Brotman RM. Translating the vaginal microbiome: gaps and challenges. Genome Med 2016;8:35. [Crossref] [PubMed]
- Godoy-Vitorino F, Romaguera J, Zhao C, et al. Cervicovaginal Fungi and Bacteria Associated With Cervical Intraepithelial Neoplasia and High-Risk Human Papillomavirus Infections in a Hispanic Population. Front Microbiol 2018;9:2533. [Crossref] [PubMed]
- Beamer MA, Austin MN, Avolia HA, et al. Bacterial species colonizing the vagina of healthy women are not associated with race. Anaerobe 2017;45:40-3. [Crossref] [PubMed]
- Brotman RM, Shardell MD, Gajer P, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis 2014;210:1723-33. [Crossref] [PubMed]
- Norenhag J, Du J, Olovsson M, et al. The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG 2020;127:171-80. [Crossref] [PubMed]
- Onywera H, Williamson AL, Mbulawa ZZA, et al. Factors associated with the composition and diversity of the cervical microbiota of reproductive-age Black South African women: a retrospective cross-sectional study. PeerJ 2019;7:e7488. [Crossref] [PubMed]
- Chen Y, Hong Z, Wang W, et al. Association between the vaginal microbiome and high-risk human papillomavirus infection in pregnant Chinese women. BMC Infect Dis 2019;19:677. [Crossref] [PubMed]
- Oh HY, Kim BS, Seo SS, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect 2015;21:674.e1-9. [Crossref] [PubMed]
- Wiik J, Sengpiel V, Kyrgiou M, et al. Cervical microbiota in women with cervical intra-epithelial neoplasia, prior to and after local excisional treatment, a Norwegian cohort study. BMC Womens Health 2019;19:30. [Crossref] [PubMed]
- Di Paola M, Sani C, Clemente AM, et al. Characterization of cervico-vaginal microbiota in women developing persistent high-risk Human Papillomavirus infection. Sci Rep 2017;7:10200. [Crossref] [PubMed]
- Borgogna JC, Shardell MD, Santori EK, et al. The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG 2020;127:182-92. [Crossref] [PubMed]
- Kyrgiou M, Mitra A, Moscicki AB. Does the vaginal microbiota play a role in the development of cervical cancer? Transl Res 2017;179:168-82. [Crossref] [PubMed]
- Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 2015;5:16865. [Crossref] [PubMed]
- Laniewski P, Cui H, Roe DJ, et al. Features of the cervicovaginal microenvironment drive cancer biomarker signatures in patients across cervical carcinogenesis. Sci Rep 2019;9:7333. [Crossref] [PubMed]
- Ilhan ZE, Laniewski P, Thomas N, et al. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine 2019;44:675-90. [Crossref] [PubMed]
- Motevaseli E, Shirzad M, Akrami SM, et al. Normal and tumour cervical cells respond differently to vaginal lactobacilli, independent of pH and lactate. J Med Microbiol 2013;62:1065-72. [Crossref] [PubMed]
Cite this article as: McKee KS, Carter KA, Bassis C, Young VB, Reed B, Harper DM, Ruffin MT 4th, Bell JD. The vaginal microbiota, high-risk human papillomavirus infection, and cervical cytology: results from a population-based study. Gynecol Pelvic Med 2020;3:18.


